Jeffrey P. Henderson, MD, PhD, FIDSA, Professor of Medicine and Molecular Microbiology, at WashU Division of Infectious Diseases, was recently published in ASM Journals with new insights into the development of a metabolome-based respiratory infection prognostic during COVID-19.
Henderson highlights that the supportive environment at WashU Medicine played a crucial role in making this work possible, an aspect that is not explicitly mentioned in the article. Contributors include: John I. Robinson, Laura R. Marks, MD, Jane A. O’Halloran, MD, WashU Medicine Division of Infectious Diseases. Along with: Charles W. Goss, PhD, Director, WashU Center for Biostatistics and Data Science, Andrew Hinton and Peter J. Mucha, Curriculum in Bioinformatics and Computational Biology, University of North Carolina, Chapel Hill.
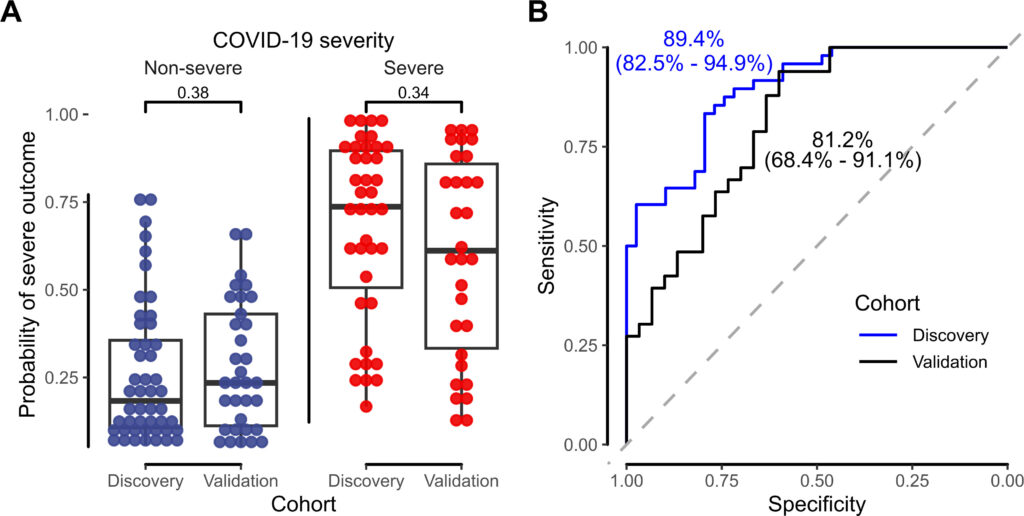
In a new respiratory virus pandemic, the ability to identify patients at greatest risk for severe disease is critical to administering antiviral therapies early, when they are most likely to be effective. While risk prediction tools are best developed early in a pandemic, availability of laboratory-based resources to develop these may be limited by available technology and by infection precautions.
“As we faced large numbers of COVID cases in early 2020 it became rapidly evident that a major challenge would be directing resources to patients who were most likely to benefit from them. Antiviral therapies and antiviral drugs in particular work best in most patients when given early. We established this interdisciplinary team in the Infectious Diseases Division to repurpose a metabolomic approach in development in my group to identify patients at high risk for severe COVID-19. Key to our success was our team’s interdisciplinary expertise in metabolomics, patient-oriented research, and clinical medicine.”
Jeffrey P. Henderson, MD, PhD, FIDSA
Here (figure 2), we show that an accessible metabolic profiling approach could identify a prognostic signature of severe disease in the initial wave of COVID-19, when patients presenting for care often exceeded the available doses of convalescent plasma and remdesivir. In a future pandemic, this approach, alongside efforts to identify clinical disease severity predictors, could improve patient outcomes and facilitate therapeutic trials by identifying individuals at high risk for severe disease.